
Biochemistry and Molecular Biophysics
Lab Website
Graduate Programs (DBBS)
Publications (PubMed / NIH)
Research
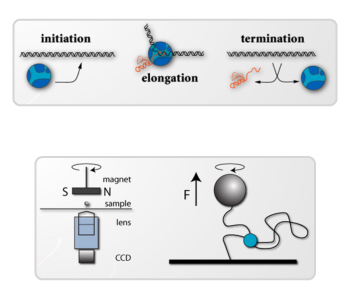
The Galburt lab studies the molecular mechanisms underlying transcription initiation and DNA repair in Bacteria and Eukaryotes. Four current lines of investigation are ongoing in the lab:
- Transcriptional regulation in Mycobacterium tuberculosis
- DNA repair helicase activation in Mycobacterium tuberculosis
- Development and application of high-throughput in vitro measures of transcription rates.
- DNA unwinding and start-site scanning by the Pre-Initiation Complex in Yeast and Humans
The lab specializes in using biochemical and biophysical techniques to monitor reactions in real-time. These techniques allow for the quantification of the rates and magnitudes of conformational transitions in RNA polymerase, its associated transcription factors, DNA helicases, and the DNA templates on which these molecular motors act. Our goal is to understand how reaction kinetics are dictated by molecular interactions and how they encode basal and regulatory biochemical activities.
Select Publications
Upasana L. Mallimadugula & Eric A. Galburt (2022). “Parallel path mechanisms lead to nonmonotonic force-velocity curves and an optimum load for molecular motor function” Phys Rev E. 2022 Mar;105(3-1):034405. doi: 10.1103/PhysRevE.105.034405. (Abstract)
Drake Jensen & Eric A. Galburt (2021). “The Context-Dependent Influence of Promoter Sequence Motifs on Transcription Initiation Kinetics and Regulation” J Bacteriol. 2021 Mar 23;203(8):e00512-20. doi: 10.1128/JB.00512-20. Print 2021 Mar 23. (Abstract)
Zhu D.X., Garner A.L., Galburt E.A., & Stallings C.L. (2019). “CarD contributes to diverse gene expression outcomes throughout the genome of Mycobacterium tuberculosis” Proc Natl Acad Sci U S A. 2019 Jul 2;116(27):13573-13581. doi: 10.1073/pnas.1900176116. Epub 2019 Jun 19. (Abstract)
Jensen D., Ruiz Manzano A., Rammohan J., Stallings C.L., & Galburt E.A. (2019). “CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase.” Nucleic Acids Res. 2019 May 25. pii: gkz449. doi: 10.1093/nar/gkz449. [Epub ahead of print] (Abstract)
Tomko E.J. & Galburt E.A. (2019). “Single-molecule approach for studying RNAP II transcription initiation using magnetic tweezers.” Methods. 2019 Mar 18. pii: S1046-2023(18)30297-4. doi: 10.1016/j.ymeth.2019.03.010. [Epub ahead of print] (Abstract)
Drake Jensen, Ana Ruiz Manzano, Christina L. Stallings, & Eric A. Galburt. (2019). “Regulation of Mycobacterial RNA Polymerase Promoter Escape Kinetics by Transcription Factors Card and RbpA” Biophysical Journal. Volume 116, Issue 3, Supplement 1, 15 February 2019, Page 210a. (Abstract)
Eric A. Galburt, Eric J. Tomko, James Fishburn, & Steven Hahn. (2019). “Tfiih Generates a Six-Base-Pair Open Complex during Eukaryotic Transcription Initiation” Biophysical Journal. Volume 116, Issue 3, Supplement 1, 15 February 2019, Pages 209a-210a. (Abstract)
Andrea Soranno, Jeremias Incicco, Paolo De Bona, Eric Tomko, Eric Galburt, & Roberto Galletto. (2019). “Shelterin Components Modulate the Phase-Separation Propensity of Telomeres” Biophysical Journal. Volume 116, Issue 3, Supplement 1, 15 February 2019, Pages 467a-468a. (Abstract)
Galburt E.A. (2018). “The calculation of transcript flux ratios reveals single regulatory mechanisms capable of activation and repression.” Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11604-E11613. doi: 10.1073/pnas.1809454115. Epub 2018 Nov 21. (Abstract)
Prusa J., Jensen D., Santiago-Collazo G., Pope S.S., Garner A.L., Miller J.J., Ruiz Manzano A., Galburt E.A., & Stallings C.L. (2018). “Domains within RbpA serve specific functional roles that regulate the expression of distinct mycobacterial gene subsets.” J Bacteriol. 2018 Apr 23. pii: JB.00690-17. doi: 10.1128/JB.00690-17. (Abstract)
Tomko E.J., Fishburn J., Hahn S., & Galburt E.A. (2017). “TFIIH generates a six-base-pair open complex during RNAP II transcription initiation and start-site scanning.” Nat Struct Mol Biol. 2017 Dec;24(12):1139-1145. doi: 10.1038/nsmb.3500. Epub 2017 Nov 6. (Abstract)
Galburt E.A. & Tomko E.J. (2017). “Conformational selection and induced fit as a useful framework for molecular motor mechanisms.” Biophys Chem. 2017 Apr;223:11-16. doi: 10.1016/j.bpc.2017.01.004. Epub 2017 Feb 3. (Abstract)
Garner A.L., Rammohan J., Huynh J.P., Onder L.M., Chen J., Bae B., Jensen D., Weiss L.A., Ruiz Manzano A., Darst S.A., Campbell E.A., Nickels B.E., Galburt E.A., & Stallings C.L. (2016). “Effects of Increasing the Affinity of CarD for RNA Polymerase on Mycobacterium tuberculosis Growth, rRNA Transcription, and Virulence.” J Bacteriol. 2017 Jan 30;199(4). pii: e00698-16. doi: 10.1128/JB.00698-16. Print 2017 Feb 15. (Abstract)
Galburt E.A. & Rammohan J. (2016). “A Kinetic Signature for Parallel Pathways: Conformational Selection and Induced Fit. Links and Disconnects between Observed Relaxation Rates and Fractional Equilibrium Flux under Pseudo-First-Order Conditions.” Biochemistry. 2016 Dec 20;55(50):7014-7022. Epub 2016 Dec 8. (Abstract)
Rammohan J., Ruiz Manzano A., Garner A.L., Prusa J. Stallings C.L., & Galburt E.A. (2016). “Cooperative stabilization of Mycobacterium tuberculosis rrnAP3 promoter open complexes by RbpA and CarD.” Nucleic Acids Res. 2016 Sep 6;44(15):7304-13. doi: 10.1093/nar/gkw577. Epub 2016 Jun 24. (Abstract)
