Current Lab Members and Collaborators:
Dr. Carl
Frieden, Dr. Jenny
Buzan, Dr. Clay Clark, Dr. Tim Close, Dr.
George Drysdale, Jinyan (Lee)
Du, Dr. Sydney
Hoeltzli, Dr. Brad Karon, Dr.
Keehyuk Kim, Ran Kim, and Dr. Linda
Kurz
You are visitor number

since 3/97
since 3/97
KINSIM and FITSIM Downloads
You can download the newest versions of Kinsim and
Fitsim for the PC
from
this site. Click Here to view the release
notes and download the compressed file. You will also find a link to a
short help page. Another PC program, KSIM, is also described on this page.
Overview of Research Projects
 |
 |
 |
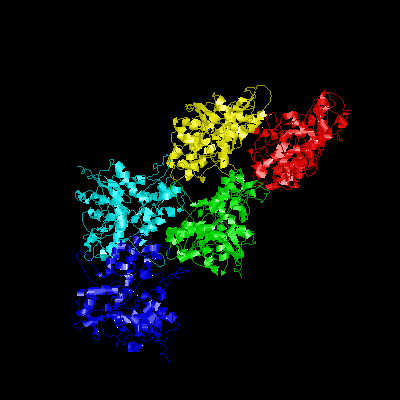 |
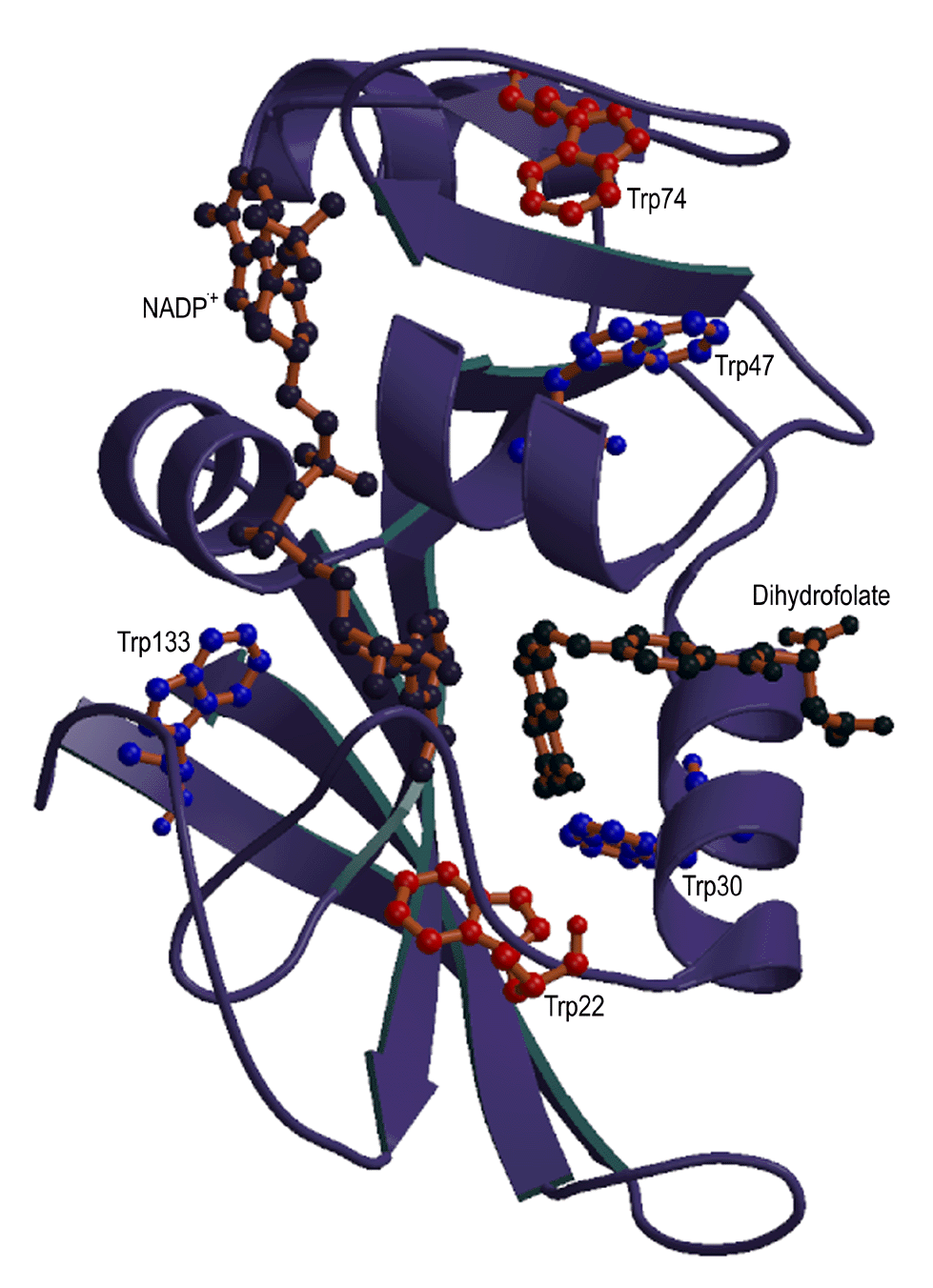
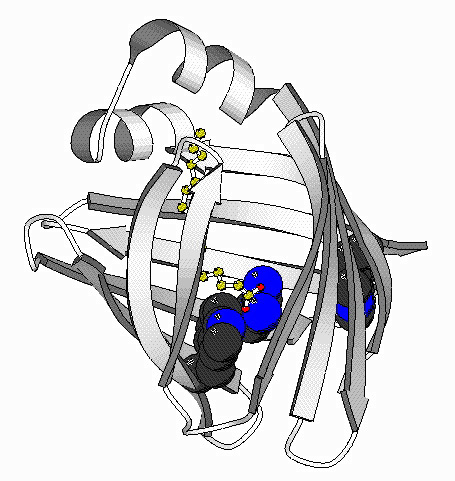
| GroEL | DHFR | IFABP | Actin |
Protein folding, protein structure/function relationships, protein-protein interactions and the
mechanism of enzymatic reactions are projects currently under study in this laboratory.
The long term goal of the protein folding studies is to understand the nature of the intermediate structures on the unfolding and refolding pathways, including the role of proteins that assist folding (called chaperonins). The work uses site-directed mutagenesis and techniques such as 19F and proton NMR, circular dichroism, fluorescence measurements and x-ray crystallography. Proteins in these studies include the intestinal fatty acid binding protein, dihydrofolate reductase, actin, and GroEL.
Studies on protein-protein interactions are related to the folding studies but also include actin, actin binding proteins and polymerization processes. The long term goal of this work is to understand the role of actin in maintaining the cytoskeletal structure of cells and how such structures control cellular functions. Studies on site-directed mutants of actin are underway.
The long term goal of kinetic studies with enzymes is to understand the catalytic mechanism of specific enzymatic reactions.
For a List of References on These and Other
Projects Click
Here
Specific Research Projects
Interactions of the Molecular Chaperonin GroEL with
Dihydrofolate Reductase