
Biochemistry and Molecular Biophysics
Lab Website
Graduate Programs (DBBS)
Publications (PubMed / NIH)
Research
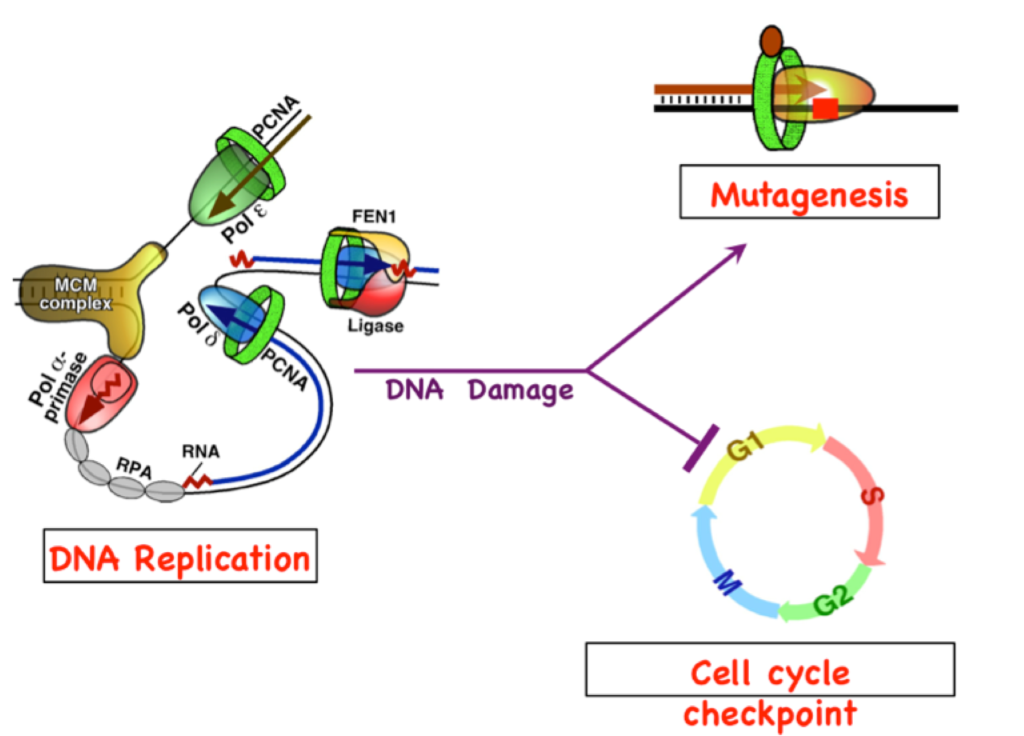
DNA replication fork and Okazaki fragment maturation
Select Publications
Das M, Hile SE, Brewster J, Boer JL, Bezalel-Buch R, Guo Q, Yang W, Burgers PM, Eckert KA, Freudenreich CH. “DNA polymerase zeta can efficiently replicate structures formed by AT/TA repeat sequences and prevent their deletion. ” Nucleic Acids Res. 2024 Dec 27:gkae1254. doi: 10.1093/nar/gkae1254. Online ahead of print. PMID: 39727171(Abstract)
Elias A. Tannous & Peter M. Burgers (2021). “Novel insights into the mechanism of cell cycle kinases Mec1(ATR) and Tel1(ATM)” Crit Rev Biochem Mol Biol. 2021 Jun 20;1-14. doi: 10.1080/10409238.2021.1925218. Online ahead of print. (Abstract)
Denis A. Kiktev, Margaret Dominska, Tony Zhang, Joseph Dahl, Elena I. Stepchenkova, Piotr Mieczkowski, Peter M. Burgers, Scott Lujan, Adam Burkholder, Thomas A. Kunkel, & Thomas D. Petes (2021). “The fidelity of DNA replication, particularly on GC-rich templates, is reduced by defects of the Fe-S cluster in DNA polymerase δ” Nucleic Acids Res. 2021 Jun 4;49(10):5623-5636. doi: 10.1093/nar/gkab371. (Abstract)
Elias A. Tannous, Luke A. Yates, Xiaodong Zhang, & Peter M. Burgers (2021). “Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase” Nat Struct Mol Biol. 2021 Jan;28(1):50-61. doi: 10.1038/s41594-020-00522-0. Epub 2020 Nov 9. (Abstract)
Melanie A. Sparks, Peter M. Burgers, & Roberto Galletto (2020). “Pif1, RPA and FEN1 modulate the ability of DNA polymerase δ to overcome protein barriers during DNA synthesis” J Biol Chem. 2020 Nov 20;295(47):15883-15891. doi: 10.1074/jbc.RA120.015699. Epub 2020 Sep 10. (Abstract)
Rachel Bezalel-Buch, Young K. Cheun, Upasana Roy, Orlando D. Schärer, & Peter M. Burgers (2020). “Bypass of DNA interstrand crosslinks by a Rev1-DNA polymerase ζ complex” Nucleic Acids Res. 2020 Jul 7;gkaa580. doi: 10.1093/nar/gkaa580. Online ahead of print. (Abstract)
Hohl M., Mojumdar A., Hailemariam S., Kuryavyi V., Ghisays F., Sorenson K., Chang M., Taylor B.S., Patel D.J., Burgers P.M., Cobb J.A., & Petrini J.H.J. (2020). “Modeling cancer genomic data in yeast reveals selection against ATM function during tumorigenesis.” PLoS Genet. 2020 Mar 18;16(3):e1008422. doi: 10.1371/journal.pgen.1008422. [Epub ahead of print] (Abstract)
Yates L.A., Williams R.M., Hailemariam S., Ayala R., Burgers P., & Zhang X. (2019). “Cryo-EM Structure of Nucleotide-Bound Tel1ATM Unravels the Molecular Basis of Inhibition and Structural Rationale for Disease-Associated Mutations.” Structure. 2019 Nov 4. pii: S0969-2126(19)30353-3. doi: 10.1016/j.str.2019.10.012. [Epub ahead of print] (Abstract)
Sparks J.L., Gerik K.J., Stith C.M., Yoder B.L., & Burgers P.M. (2019). “The roles of fission yeast exonuclease 5 in nuclear and mitochondrial genome stability.” DNA Repair (Amst). 2019 Nov;83:102720. doi: 10.1016/j.dnarep.2019.102720. Epub 2019 Sep 21. (Abstract)
Hailemariam S., De Bona P., Galletto R., Hohl M., Petrini J.H., & Burgers P.M. (2019). “The telomere-binding protein Rif2 and ATP-bound Rad50 have opposing roles in the activation of yeast Tel1ATM kinase.” J Biol Chem. 2019 Oct 22. pii: jbc.RA119.011077. doi: 10.1074/jbc.RA119.011077. [Epub ahead of print] (Abstract)
Sparks M.A., Singh S.P, Burgers P.M., & Galletto R. (2019). “Complementary roles of Pif1 helicase and single stranded DNA binding proteins in stimulating DNA replication through G-quadruplexes.” Nucleic Acids Res. 2019 Jul 24. pii: gkz608. doi: 10.1093/nar/gkz608. [Epub ahead of print] (Abstract)
Sarem Hailemariam, Sandeep Kumar, & Peter M. Burgers (2019). “Activation of Tel1ATM kinase requires Rad50 ATPase and long nucleosome-free DNA, but no DNA ends.” J Biol Chem. 2019 Jun 28;294(26):10120-10130. doi: 10.1074/jbc.RA119.008410. Epub 2019 May 9. (Abstract)
Mondol T., Stodola J.L., Galletto R., & Burgers P.M. (2019). “PCNA accelerates the nucleotide incorporation rate by DNA polymerase δ.” Nucleic Acids Res. 2019 Feb 28;47(4):1977-1986. doi: 10.1093/nar/gky1321. (Abstract)
Burgers, P.M. (2019). “Solution to the 50-year-old Okazaki-fragment problem.” Proc Natl Acad Sci U S A. 2019 Feb 26;116(9):3358-3360. doi: 10.1073/pnas.1900372116. Epub 2019 Feb 15. (Abstract)
Stodola J.L. & Burgers P.M. (2017). “Mechanism of Lagging-Strand DNA Replication in Eukaryotes.” Adv Exp Med Biol. 2017;1042:117-133. doi: 10.1007/978-981-10-6955-0_6. (Abstract)
