
Professor
Biochemistry and Molecular Biophysics
Lab Website
Graduate Programs (DBBS)
Publications (PubMed / NIH)
Research
Mechanistic studies of DNA motor proteins.
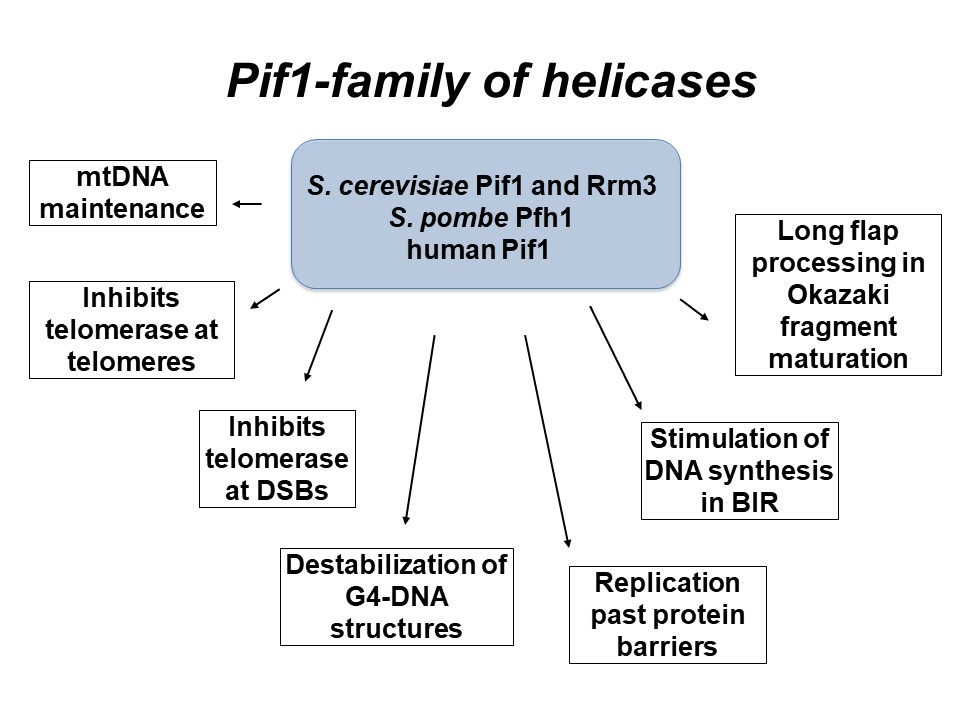
Select Publications
Mor Varon, Daniel Dovrat, Jonathan Heuzé, Ioannis Tsirkas, Saurabh P Singh, Philippe Pasero, Roberto Galletto, & Amir Aharoni (2024). “Rrm3 and Pif1 division of labor during replication through leading and lagging strand G-quadruplex” Nucleic Acids Res. 2024 Feb 28;52(4):1753-1762. doi: 10.1093/nar/gkad1205. (Abstract)
Melanie A. Sparks, Peter M. Burgers, & Roberto Galletto (2020). “Pif1, RPA and FEN1 modulate the ability of DNA polymerase δ to overcome protein barriers during DNA synthesis” J Biol Chem. 2020 Nov 20;295(47):15883-15891. doi: 10.1074/jbc.RA120.015699. Epub 2020 Sep 10. (Abstract)
Singh S.P., Soranno A., Sparks M.A., & Galletto R. (2019). “Branched unwinding mechanism of the Pif1 family of DNA helicases.” Proc Natl Acad Sci U S A. 2019 Nov 19. pii: 201915654. doi: 10.1073/pnas.1915654116. [Epub ahead of print] (Abstract)
Hailemariam S., De Bona P., Galletto R., Hohl M., Petrini J.H., & Burgers P.M. (2019). “The telomere-binding protein Rif2 and ATP-bound Rad50 have opposing roles in the activation of yeast Tel1ATM kinase.” J Biol Chem. 2019 Oct 22. pii: jbc.RA119.011077. doi: 10.1074/jbc.RA119.011077. [Epub ahead of print] (Abstract)
Sparks M.A., Singh S.P, Burgers P.M., & Galletto R. (2019). “Complementary roles of Pif1 helicase and single stranded DNA binding proteins in stimulating DNA replication through G-quadruplexes.” Nucleic Acids Res. 2019 Jul 24. pii: gkz608. doi: 10.1093/nar/gkz608. [Epub ahead of print] (Abstract)
Singh S.P., Kukshal V., & Galletto R. (2019). “A stable tetramer is not the only oligomeric state that mitochondrial single-stranded DNA binding proteins can adopt.” J Biol Chem. 2019 Mar 15;294(11):4137-4144. doi: 10.1074/jbc.RA118.007048. Epub 2019 Jan 7. (Abstract)
Mondol T., Stodola J.L., Galletto R., & Burgers P.M. (2019). “PCNA accelerates the nucleotide incorporation rate by DNA polymerase δ.” Nucleic Acids Res. 2019 Feb 28;47(4):1977-1986. doi: 10.1093/nar/gky1321. (Abstract)
Andrea Soranno, Jeremias Incicco, Paolo De Bona, Eric Tomko, Eric Galburt, & Roberto Galletto. (2019). “Shelterin Components Modulate the Phase-Separation Propensity of Telomeres” Biophysical Journal. Volume 116, Issue 3, Supplement 1, 15 February 2019, Pages 467a-468a. (Abstract)
Dahan D., Tsirkas I., Dovrat D., Sparks M.A., Singh S.P., Galletto R., & Aharoni A. (2018). “Pif1 is essential for efficient replisome progression through lagging strand G-quadruplex DNA secondary structures.” Nucleic Acids Res. 2018 Dec 14;46(22):11847-11857. doi: 10.1093/nar/gky1065. (Abstract)
Geronimo C.L., Singh S.P., Galletto R., & Zakian V.A. (2018). “The signature motif of the Saccharomyces cerevisiae Pif1 DNA helicase is essential in vivo for mitochondrial and nuclear functions and in vitro for ATPase activity.” Nucleic Acids Res. 2018 Sep 19;46(16):8357-8370. doi: 10.1093/nar/gky655. (Abstract)
Singh S.P., Kukshal V., De Bona P., Antony E., & Galletto R. (2018). “The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.” Nucleic Acids Res. 2018 Aug 21;46(14):7193-7205. doi: 10.1093/nar/gky530. (Abstract)
Sokoloski J.E., Kozlov A.G., Galletto R., & Lohman T.M. (2016). “Chemo-mechanical pushing of proteins along single-stranded DNA.” Proc Natl Acad Sci U S A. 2016 May 31;113(22):6194-9. doi: 10.1073/pnas.1602878113. Epub 2016 May 16. (Abstract)
Koc K.N., Singh S.P., Stodola J.L., Burgers P.M., & Galletto R. (2016). “Pif1 removes a Rap1-dependent barrier to the strand displacement activity of DNA polymerase δ.” Nucleic Acids Res. 2016 May 5;44(8):3811-9. doi: 10.1093/nar/gkw181. Epub 2016 Mar 21. (Abstract)
Singh, S.P., Koc, K.N., Stodola, J.L. and Galletto, R.
A Monomer of Pif1 Unwinds Double-Stranded DNA and It Is Regulated by the Nature of the Non-Translocating Strand at the 3′-End.
J Mol Biol 428:1053-1067 (2016). (Abstract)
Feldmann, E.A., De Bona, P. and Galletto, R.
The wrapping loop and Rap1 C-terminal (RCT) domain of yeast Rap1 modulate access to different DNA binding modes.
Journal of Biological Chemistry 290:11455-11466 (2015). (Abstract)
