
Biochemistry and Molecular Biophysics
Lab Website
Publications (PubMed / NIH)
Research
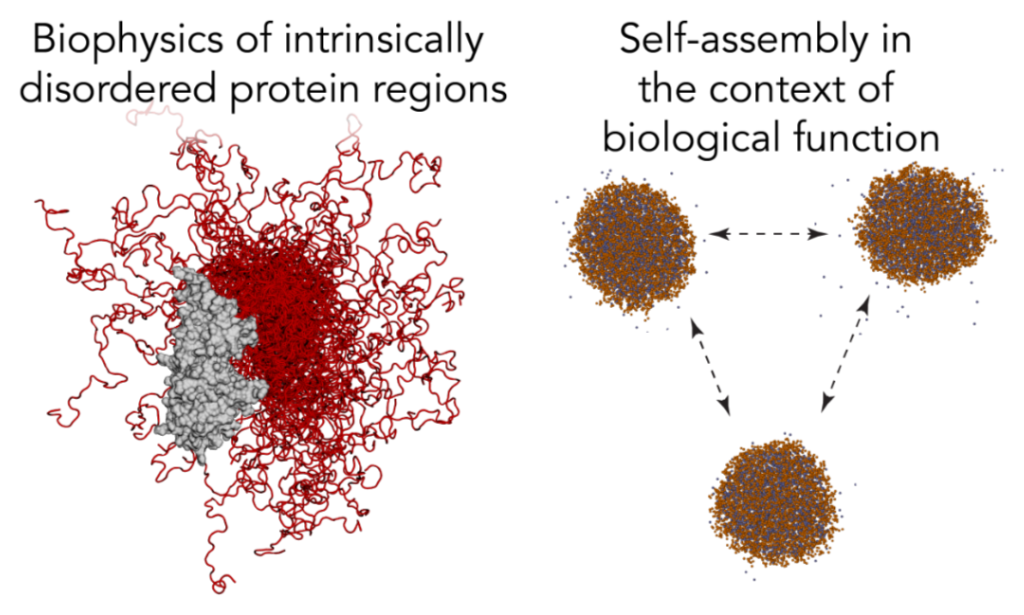
We explore how intrinsically disordered protein regions confer biological function and how this goes wrong in disease. The lab integrates physics-based models (all-atom and coarse-grained simulations) with informatics and machine learning to develop sequence-specific predictions. We then test those predictions either within the lab, or with collaborators around the world.
Of particular interest is understanding how disordered proteins drive self-assembly in the context of biological phase separation, with a major emphasis on understanding when, how, and why this might be important for normal cellular function.
Select Publications
Meaghan S Jankowski, Daniel Griffith, Divya G Shastry, Jacqueline F Pelham, Garrett M Ginell, Joshua Thomas, Pankaj Karande, Alex S Holehouse & Jennifer M Hurley (2024). “Disordered clock protein interactions and charge blocks turn an hourglass into a persistent circadian oscillator” Nat Communn. 2024 Apr 25;15(1):3523. doi: 10.1038/s41467-024-47761-z. (Abstract)
Zongru Li, Qionghua Shen, Emery T Usher, Andrew P Anderson, Manuel Iburg, Richard Lin, Brandon Zimmer, Matthew D Meyer, Alex S Holehouse, Lingchong You, Ashutosh Chilkoti, Yifan Dai & George J Lu (2024). “Phase transition of GvpU regulates gas vesicle clustering in bacteria” Nat Microbiol. 2024 Apr;9(4):1021-1035. doi: 10.1038/s41564-024-01648-3. Epub 2024 Mar 29. (Abstract)
S Sanchez-Martinez, K Nguyen, S Biswas, V Nicholson, A V Romanyuk, J Ramirez, S Kc, A Akter, C Childs, E K Meese, E T Usher, G M Ginell, F Yu, E Gollub, M Malferrari, F Francia, G Venturoli, E W Martin, F Caporaletti, G Giubertoni, S Woutersen, S Sukenik, D N Woolfson, A S Holehouse & T C Boothby (2024). “Labile assembly of a tardigrade protein induces biostasis” Protein Sci.2024 Apr;33(4):e4941. doi: 10.1002/pro.4941. (Abstract)
Jeffrey M Lotthammer, Garrett M Ginell, Daniel Griffith, Ryan J Emenecker & Alex S Holehouse (2024). “Direct prediction of intrinsically disordered protein conformational properties from sequence” Nat Methods. 2024 Mar;21(3):465-476. doi: 10.1038/s41592-023-02159-5. Epub 2024 Jan 31. (Abstract)
Sourav Biswas, Edith Gollub, Feng Yu, Garrett Ginell, Alex Holehouse, Shahar Sukenik & Thomas C Boothby 1 (2024). “Helicity of a tardigrade disordered protein contributes to its protective function during desiccation” Protein Sci. 2024 Feb;33(2):e4872. doi: 10.1002/pro.4872. (Abstract)
David Moses, Karina Guadalupe, Feng Yu, Eduardo Flores, Anthony R Perez, Ralph McAnelly, Nora M Shamoon, Gagandeep Kaur, Estefania Cuevas-Zepeda, Andrea D Merg, Erik W Martin, Alex S Holehouse & Shahar Sukenik (2024). “Structural biases in disordered proteins are prevalent in the cell” Nat Struct Mol Biol. 2024 Feb;31(2):283-292. doi: 10.1038/s41594-023-01148-8. Epub 2024 Jan 4. (Abstract)
Alex S Holehouse & Birthe B Kragelund (2024). “The molecular basis for cellular function of intrinsically disordered protein regions” Nat Rev Mol Cell Biol. 2024 Mar;25(3):187-211. doi: 10.1038/s41580-023-00673-0. Epub 2023 Nov 13. (Abstract)
Priya R Banerjee, Alex S Holehouse, Richard Kriwacki, Paul Robustelli, Hao Jiang, Alexander I Sobolevsky, Jennifer M Hurley & Joshua T Mendell (2024). “Dissecting the biophysics and biology of intrinsically disordered proteins” Trends Biochem Sci. 2024 Feb;49(2):101-104. doi: 10.1016/j.tibs.2023.10.002. Epub 2023 Nov 8. (Abstract)
David Moses, Garrett M Ginell, Alex S Holehouse & Shahar Sukenik (2023). “Intrinsically disordered regions are poised to act as sensors of cellular chemistry” Trends Biochem Sci. 2023 Dec;48(12):1019-1034. doi: 10.1016/j.tibs.2023.08.001. Epub 2023 Aug 31. (Abstract)
Karol Buda, Katerina Cermakova, H Courtney Hodges, Eugenio F Fornasiero, Shahar Sukenik & Alex S Holehouse (2023). “Using graphs and charts in scientific figures” Trends Biochem Sci. 2023 Nov;48(11):913-916. doi: 10.1016/j.tibs.2023.08.011. (Abstract)
Jasmine Cubuk, Lina Greenberg, Akiva E Greenberg, Ryan J Emenecker, Melissa D Stuchell-Brereton, Alex S Holehouse, Andrea Soranno & Michael J Greenberg (2024). “Structural dynamics of the intrinsically disordered linker region of cardiac troponin T” bioRxiv. [Preprint]. 2024 May 31:2024.05.30.596451. doi: 10.1101/2024.05.30.596451. (Abstract)
Jasmine Cubuk, Jhullian J Alston, J Jeremías Incicco, Alex S Holehouse, Kathleen B Hall, Melissa D Stuchell-Brereton & Andrea Soranno (2024). “The disordered N-terminal tail of SARS-CoV-2 Nucleocapsid protein forms a dynamic complex with RNA” Nucleic Acids Res. 2024 Mar 21;52(5):2609-2624. doi: 10.1093/nar/gkad1215. (Abstract)
Garrett M. Ginell & Alex S. Holehouse (2022). “An Introduction to the Stickers-and-Spacers Framework as Applied to Biomolecular Condensates” Methods Mol Biol. 2023;2563:95-116. doi: 10.1007/978-1-0716-2663-4_4. (Abstract)
Nicolás S. González-Foutel, Juliana Glavina, Wade M. Borcherds, Matías Safranchik, Susana Barrera-Vilarmau, Amin Sagar, Alejandro Estaña, Amelie Barozet, Nicolás A. Garrone, Gregorio Fernandez-Ballester, Clara Blanes-Mira, Ignacio E. Sánchez, Gonzalo de Prat-Gay, Juan Cortés, Pau Bernadó, Rohit V. Pappu, Alex S. Holehouse, Gary W. Daughdrill, & Lucía B. Chemes (2022). “Conformational buffering underlies functional selection in intrinsically disordered protein regions” Nat Struct Mol Biol. 2022 Aug;29(8):781-790. doi: 10.1038/s41594-022-00811-w. Epub 2022 Aug 10. (Abstract)
Daniel Griffith & Alex S. Holehouse (2021). “PARROT is a flexible recurrent neural network framework for analysis of large protein datasets” Elife. 2021 Sep 17;10:e70576. doi: 10.7554/eLife.70576. (Abstract)
